

Document Type : Original Article
Authors
1 Department of Animal Science, Faculty of Agriculture, Ferdowsi University of Mashhad, Iran
2 Department of Animal Science, Faculty of Agriculture, University of Aleppo, Syria
Abstract
Keywords
The contribution of sheep (Ovis aries) as a source of meat, wool, skin, and milk in Iran and worldwide is undeniable. The studies of genetic characteristics for different sheep breeds provide the guiding information for breeding and conservation programs to maintain and improve production efficiency. Mitochondrial DNA has previously shown the good potential of use in genetic and population evolution studies [1]. Iran is considered the primary sheep domestication place in the world with high sheep numbers (50 million) and diversity in breeds and ecotypes (27 different breeds and ecotypes) [2]. Sheep domestication initially took place in the broad area north of the Zagros mountains range of Iran to the southeast of Anatolia in Turkey. A geographical connection between Asia and Europe was found due to the regions of Asia Minor and the Middle East, which helped the gene flow, mixing, and differentiation since sheep domestication to happen [3]. There are already over two hundred known breeds of sheep throughout the world, which is considered a diversity barely seen in other animals. Furthermore, some animal scientists believe that domesticated sheep was originated from the European and Asian wild sheep [4].
Indigenous breeds of a certain country are considered a national and strategic resource of that country’s economy and prosperity. Due to the low selection, the high number of breeders, and the limited use of artificial insemination, these breeds are often highly genetically diverse. The increasing awareness of indigenous breeds’ importance as a genetic source to introduce new breeds of desirable traits has necessitated the preservation of genetic diversity of the native livestock [5]. Furthermore, it is reported that by decreasing the genetic diversity within a species, it becomes less likely to adapt to biotic and abiotic stresses [6]. Therefore, indigenous breeds’ genetic diversity should be carefully calculated in order to perform efficient conservation that relies on a base of in-depth Genetics knowledge [7].
The mammalian mitochondrial genome is a circular double-stranded DNA molecule of about 16 Kb that contains the ribosomal, transporter, and messenger RNAs genes. In sheep, the mitochondrial genome comprises 16.58 kb with a gene layout and organize similar to those of other mammals [8].
The evolution rate of mtDNA is approximately 6 times faster than nuclear DNA [9]. Furthermore, maternal mtDNA is inherited, haploid, and non-recombinant [10]. These features rendered mtDNA as a useful tool to determine relationships between individuals within and between species [9][11] and a useful marker for cross-sectional diagnosis among groups that have been separated for 100-1000 years [12].
Iran has diverse phenotypes of sheep regarding morphology of the tail. Several studies investigated the genetic distances between some domestic breeds, although there was not a comparison between fat-tailed, thin-tailed and semi fat-tailed breeds. In light of the previous hypotheses, we tried to use the sequence of mitochondrial control region to investigate the phylogenetic relationship between some domestic sheep breeds of Iran, Middle Eastern countries and major mitochondrial haplotypes.
Three Iranian sheep breeds (Zel, Dalagh, and Mehrabani) were chosen for the study, and five blood samples were collected from each breed. These breeds were selected for this study based on their tail size and shape as presented in (Table 1) along with other breeds’ characteristics. The total DNA extraction was performed using NEXprepTM Blood DNA Mini Kit (NEX Diagnostics- South Korea). DNA quality and quantity were determined using gel electrophoresis (Gel Red stained 1% agarose) and Epoch microplate reader (BioTek, USA), respectively.
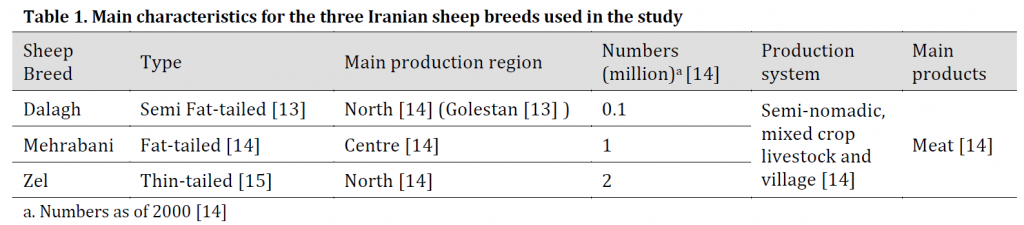
Table 1. Main characteristics for the three Iranian sheep breeds used in the study
To amplify the mitochondrial control region, a primer pair was designed according to the reference sequence (accession number NC_001941.1) (Table 2) and using Primer Premier 5 software (PREMIER Biosoft, USA). The primer pair was then evaluated with primer BLAST against the NCBI database.
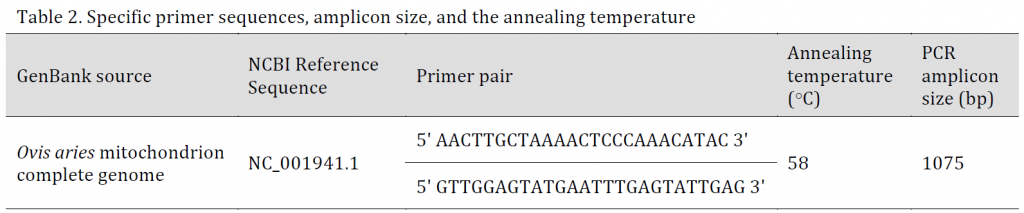
Table 2. Specific primer sequences, amplicon size, and the annealing temperature
PCR was performed using the T-Personal thermocycler (Biometra, Germany). The PCR program consisted of 10 min initial enzyme activation period at 95 ○C followed by 38 cycles of denaturation (95 ○C for 45 seconds), annealing (58 ○C for 30 seconds), and amplification (72 ○C for 90 seconds); the program was concluded with a final amplification step of 10 minutes at 72 ○C.
After electrophoresis, PCR products were analyzed by electrophoresis on 1% agarose gel and the amplified products were purified using the Gel extraction kit (Dena Zist Asia, Iran); Then, 20 μL of the purified PCR products were sent to be bi-directional sequenced. The quality of the obtained sequences was evaluated using BioEdit 5.2.7 and Chromas Lite 2.01 software.
To determine the phylogenetic relationships and overlap, all the sequences were aligned against the haplotype sequences (Haplotype A: AY829400, B: HM236176, C: DQ903302, D: HM236180, and E: HM236183) in addition to Middle Eastern sheep sequences (Turkey: Herik (KF677119), Sakiz (KF677172) and Karayaka (JN574157), Saudi Arabia: Naeimi (JN574177), Egypt: Saidi (KC461241) and Ossimi (JN573952), Iraq: Karadi (FJ545136), Hamdani (FJ545135), and Syria: Awassi (FJ545134)). The phylogenetic tree was plotted and genetic distances estimated using CLC Sequence Viewer 7.6 software (Qiagen, Germany).
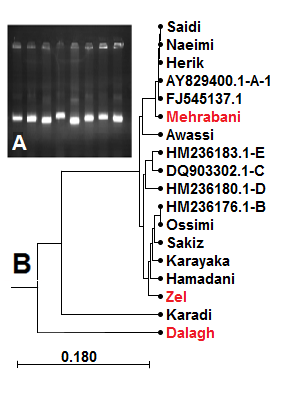
DNA was successfully extracted from all samples. The results of spectroscopy and agarose gel electrophoresis showed that the extracted DNA samples were free of contaminations and with high integrity. After PCR, electrophoresis of the reaction product on 1% agarose gel showed that the designed primers amplified the targeted template efficiently and the 1075 bp amplicons bands were sharp and specific (Fig. 1 A)
All the amplified fragments were sequenced, and the results were evaluated in terms of sequencing quality and accuracy. The low-quality nucleotides were trimmed from both ends of the target amplicons. The final sequences were recorded in the NCBI Nucleotide database under GenBank accession numbers: (Zel: MH699973, Dalag: MH699967, and Mehrabani: MH553570).
The phylogenetic tree (Fig. 1 B) showed that the Mehrabani breed was placed along with other breeds (Saeedi, Naimi, and Haryk) in haplotype A, and all of which were fat-tailed. On the other hand, Zel breed was located in Haplotype B alongside the European thin-tailed breeds, which was in agreement with other reports regarding the domestication of Iranian sheep, which illustrated that most of the Iranian sheep breeds such as Moghani, Lori Bakhtiari, Sangsari, Baluchi, Kermani, Shal, Karakul, and Kordi are located in haplotype A. [1][16][17]. Interestingly, phylogenetic results showed that Dalagh breed was close to Zel breed, probably due to their close herding locations distribution in Mazandaran and Golestan provinces, North of Iran. The Awassi breed was found in haplotype E, which is consistent with the results of [3]. Our results also showed that Iranian sheep breeds are closer to those of neighboring countries (Iraq and Syria) than those of more distant locations (Turkey, Egypt, and Saudi Arabia), which was an expected result due to geographical distances.
In a study of European, Caucasian and Asian sheep mitochondrial genome diversity, it was reported that most Asian sheep breeds were located in haplotypes B and A with 72% and 22% of the studied breeds, respectively [18].
So far, five haplotypic sheep groups have been identified (A-E) [19] where haplotypes A and B breeds are mainly located in Asia and Europe, haplotype breeds C in China, Caucasus, Turkey, and Portugal, haplotype D in the Caucasus, Karachi, and Romania, while haplotypes group E in the Middle East region.
The geographical locations of both haplotypes A and B were first reported by Wood and Phua [20]. Following, Hiendleder et al [21], carried out a complete sequencing of the domestic sheep mitochondrial genome and compared several different European and Asian breeds, and reported two major haplotype groups A and B originated from Asian and European sheep. They also suggested that the emergence of these two groups was due to the fact that present-day sheep breeds were derived from two different maternal ancestors and occurred almost simultaneously in the Near East.
Based on previous reports, most fat-tailed breeds of Iran categorized as haplotype A which is mainly originated in Asia and the Middle East. The Zel breed, the only thin tailed sheep of Iran, placed closer to haplotype B which might indicate a different domestication history. Therefore, further research is required to assess the divergence time of Iranian native sheep breeds.
Conflict of interest statement
The authors declared no conflict of interest.
Funding statement
The authors declared that this study was supported by the Ferdowsi University of Mashhad; Grant number: 3/44590.
Data availability statement
The authors declared that all related data are included in the article.